Biography
I am a AI Scientist at Paige AI. I did my Ph.D. with Jennifer Dy, Dana Brooks, and Jan-Willem van de Meent at Northeastern University. My main research interests are machine learning with emphasis on probabilistic programming, deep neural networks, and their applications in biomedical image processing. I am one of the developers of Probabilistic Torch, a library for deep generative models that extends PyTorch. I am also one of the maintainers of the Pytorch distributions module.
- Probabilistic Programming
- Deep Learning
- Biomedical Image Processing
-
PhD in Machine Learning, 2020
Northeastern University
-
MSc in Image Processing, 2013
Bilkent University
-
BSc in Electrical and Electronics Engineering, 2011
Bilkent University
Featured Publications
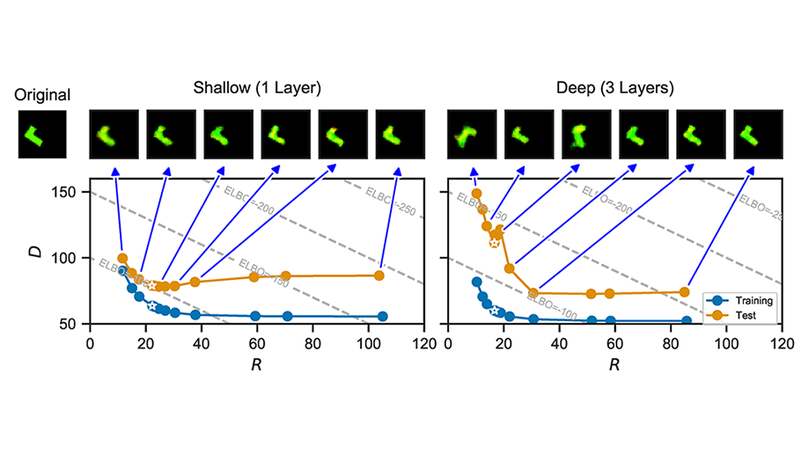
Variational autoencoders (VAEs) optimize an objective that comprises a reconstruction loss (the distortion) and a KL term (the rate). The rate is an upper bound on the mutual information, which is often interpreted as a regularizer that controls the degree of compression. We here examine whether inclusion of the rate term also improves generalization. We perform rate-distortion analyses in which we control the strength of the rate term, the network capacity, and the difficulty of the generalization problem. Lowering the strength of the rate term paradoxically improves generalization in most settings, and reducing the mutual information typically leads to underfitting. Moreover, we show that generalization performance continues to improve even after the mutual information saturates, indicating that the gap on the bound (i.e. the KL divergence relative to the inference marginal) affects generalization. This suggests that the standard spherical Gaussian prior is not an inductive bias that typically improves generalization, prompting further work to understand what choices of priors improve generalization in VAEs.
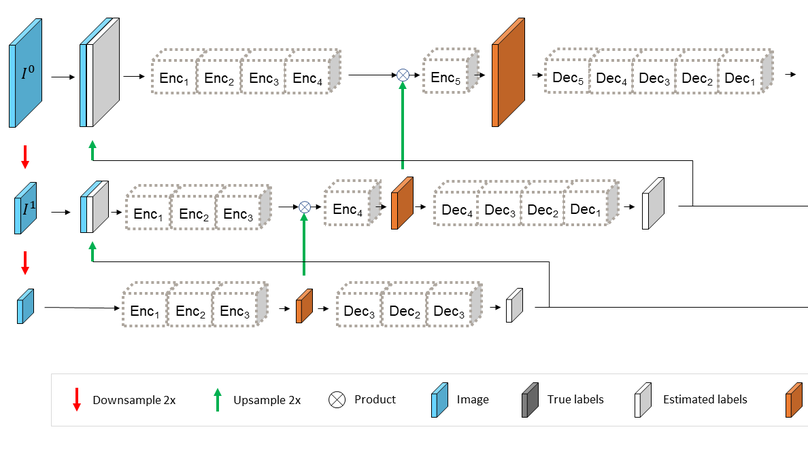
In-vivo optical microscopy is advancing into routine clinical practice for non-invasively guiding diagnosis and treatment of cancer and other diseases, and thus beginning to reduce the need for traditional biopsy. However, reading and analysis of the optical microscopic images are generally still qualitative, relying mainly on visual examination. Here we present an automated semantic segmentation method called ‘Multiscale Encoder-Decoder Network (MED-Net)’ that provides pixel-wise labeling into classes of patterns in a quantitative manner. The novelty in our approach is the modeling of textural patterns at multiple scales. This mimics the procedure for examining pathology images, which routinely starts with low magnification (low resolution, large field of view) followed by closer inspection of suspicious areas with higher magnification (higher resolution, smaller fields of view). We trained and tested our model on non-overlapping partitions of 117 reflectance confocal microscopy (RCM) mosaics of melanocytic lesions, an extensive dataset for this application, collected at four clinics in the US, and two in Italy. With patient-wise cross-validation, we achieved pixel-wise mean sensitivity and specificity of 70±11% and 95±2%, respectively, with 0.71±0.09 Dice coefficient over six classes. In the scenario, we partitioned the data clinic-wise and tested the generalizability of the model over multiple clinics. In this setting, we achieved pixel-wise mean sensitivity and specificity of 74% and 95%, respectively, with 0.75 Dice coefficient. We compared MED-Net against the state-of-the-art semantic segmentation models and achieved better quantitative segmentation performance. Our results also suggest that, due to its nested multiscale architecture, the MED-Net model annotated RCM mosaics more coherently, avoiding unrealistic-fragmented annotations.
Recent Publications
Contact
- alican@ece.neu.edu
- 805 Columbus Ave., Boston, MA 02130
- Northeastern University Interdisciplinary Science & Engineering Complex, Room 506